LENSai BioIntelligence Suite
Epitope Binning
Effortless in silico mAb Epitope Binning with proven wet lab validation
Next-level in silico epitope binning, powered by LENSai™ technology, offers a pivotal advancement with its ability to analyze over 5,000 sequences, delivering rapid insights for early triaging. This advanced method allows you, within large pools of antibodies, to detect clusters of clones that share similar target binding regions ensuring a thorough and inclusive selection process. LENSai epitope binning smart algorithms enhance biological research, offering accurate, high-throughput candidate selection while reducing time and costs.
Innovate earlier. Scale smarter.
Key benefits
Speed and High‑throughput:
Early epitope landscape profiling:
Proven:
Effortless:
Results within hours for small subsets to 2 weeks for >5k antibodies.
A critical step in antibody selection.
Epitope binning is crucial in the early stages of antibody discovery, as it facilitates the categorization of antibodies with similar target binding regions on the target. In general, insights in the epitope landscape of antibody panels significantly advance development of multi-targeting approaches. Additionally, understanding which protein regions are targeted by your antibody panel can significantly enhance intellectual property security.
Maximize early triaging with high efficiency
High precision with multi-dimensional clustering
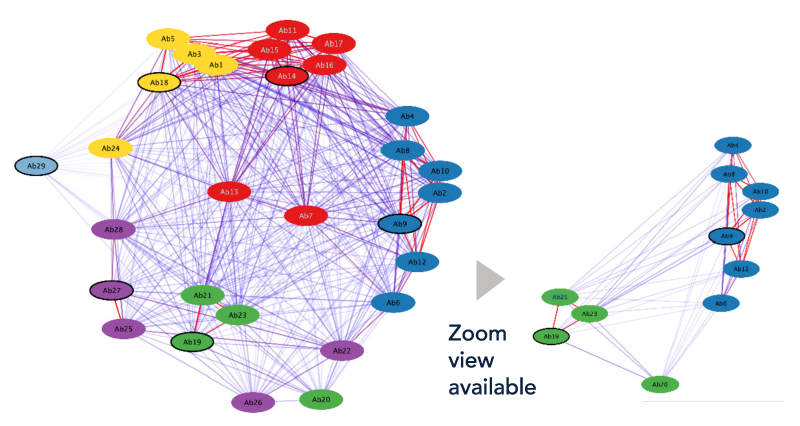
LENSai proprietary Epitope Binning algorithm includes:
- Antibody sequential and structural profiling
- Docking information, accounting for steric hindrance and glycosylation sites
- Atomic interactions of Ab-Ag complexes
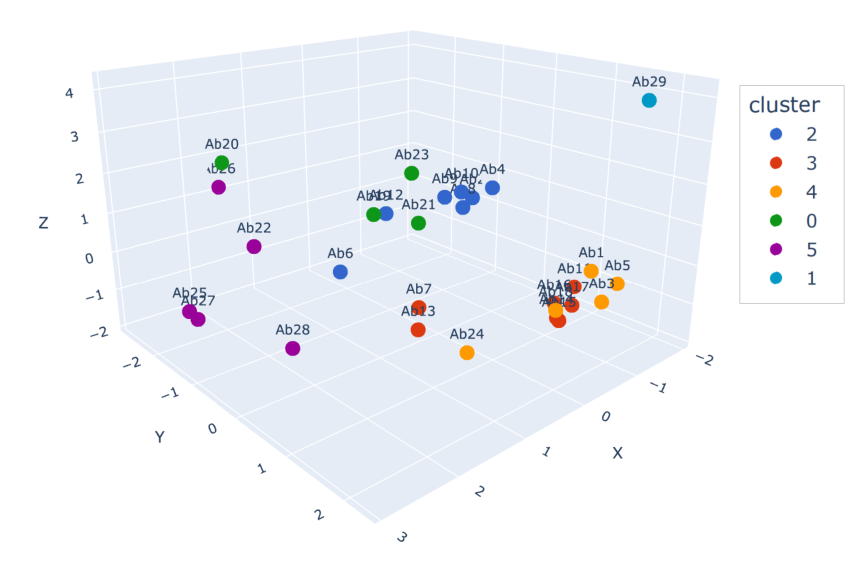
3D Grid view:
Clusters group antibodies that share a similar target binding region. Quick view on cluster proximity.
Proven results with Cohen’s Kappa testing
Blinded wet lab validation of in silico prediction of epitope bins:
- Example set of 29 antibodies
- LENSai in silico epitope prediction versus classical in vitro competition assay for binning
- Inter-annotator agreement analysis (Cohen’s Kappa Test) using a majority consensus label mapping
Case study conclusion:
Near perfect agreement between LENSai in silico Epitope Binning and classical wet-lab binning results.
- Label mapping κ
- LENSai cluster ids >
Wet lab bin 0.925 - Wet lab bin labels >
LENSai cluster ids 0.842
- Cohen's Kappa Coefficient value key:
> 0.8: Near perfect agreement